Some time ago I worked on a very important project about DNA variants analysis. Everyone in this
world has a lot of DNA variants, most of them mean nothing, but some of them are important. Scientists from different
parts of the world have opened gigabytes of useful knowledge about different variants. The goal of our project is to find
something useful in this heap of data and probably save life to a patient.

What is DNA variant?
DNA is a very complicated thing, but we as IT engineers can use an abstraction and consider it as a sequence made of symbols “A”, “C, “G” and “T”, i.e. “ACCCGTTTA…”. Everyone on our planet (at least for now) has almost the same DNA sequence, but one may have some difference in some point comparing to the majority of other people - this is called a variant.
Everyone can provide a sample to a laboratory and get a personal list of variants. Usually a laboratory provides list of variants in some part of DNA (genes) depending on its specialization. A list of variants describes positions and other properties of variants, depending on laboratory a list can be size up to several gigabytes.
What do we know about variants?
We know much about them. The problem is that this knowledge is distributed into different databases which are quite big (up to 500 gigabytes). A doctor must have opportunity to find all possible information for all variants of a patient - now the ball is in ours court to provide it.
Why not just indexing?
Unfortunately, it is not so simple. Yes, some famous variants are known as a reason of some diseases, like variant in gene X causes disease Y, so if a patient has X variant we can perform some actions. Some knowledge items are smarter, something like: “If a patient has variant X1 or variant2 together with variant Y which has frequency among Indian > 0.1” => it means a patient may have disease D”. Conditions like that make us think that we need cartesian product of VARIANT x KNOWLEDGE to solve the problem, but it is long and expensive.
Our optimization (example with house party)
We expected not too many patients per day, but we didn’t want them to wait long for result. That is why we need powerful computation equipment for each patient, but we want to optimize charges for that - this is the first point of optimization.
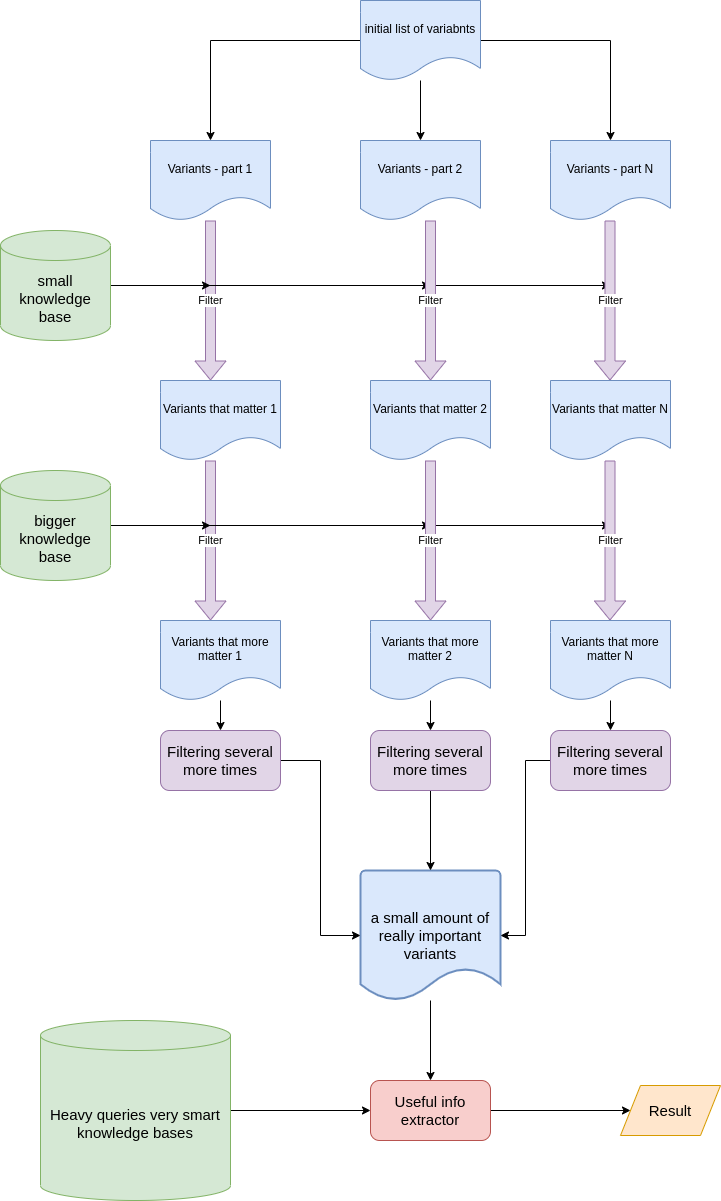
The second point is algorithm optimization. I can’t expose all secrets, but can explain the common idea. Imagine you are going to have a party at you home tonight. I’m sure you have a big house and have 1000 friends. Every friend prefer some dish/drink which you can buy in the shops of your city. You can go shopping and visit all shops and find all needed items for all your 1000 friends and provide the best party, but it is too expensive. The better option is to call your friends and understand who is going to visit you today and then provide items only for guests who confirmed the visit. Almost the same idea has been implemented in our case, we just filtered the initial list of variants by some criteria to avoid hard queries to big databases of knowledge. It was just an elegant way of using knowledge databases ony by one is a sequence, cheap first.
Prototype
We used MapReduce model to handle patient’s variations. Some variants can be handled/excluded/annotated independently of other variants that is why MapReduce model is extremely useful here. The process of handling each part of list of variations is a lot of resources consuming and needs specific soft to be used. We wanted it to be launched in some prepared environment where all soft is already installed. We were discussing using Spark or AWS Fargate for that purposes. Due to the fact, that we didn’t expect load all time, AWS Fargate has been chosen. It can be launched via API, perform some calculations and shut down after - pay only when we use it. After all parts of variants are handled, we unite them together in one list again and perform final handling that is almost cartesian product with several knowledge bases. However, it is acceptable because the amount of variants is not so huge like in the beginning.
Results
Summarizing all together, we built a prototype of the system which is able to provide invaluable information for a patient by given list of variants using the most advanced human knowledge bases. It takes several minutes and not expensive. The system is easy scalable in both sides, so you can handle high load if it comes and not pay much when you don’t have many patients.
